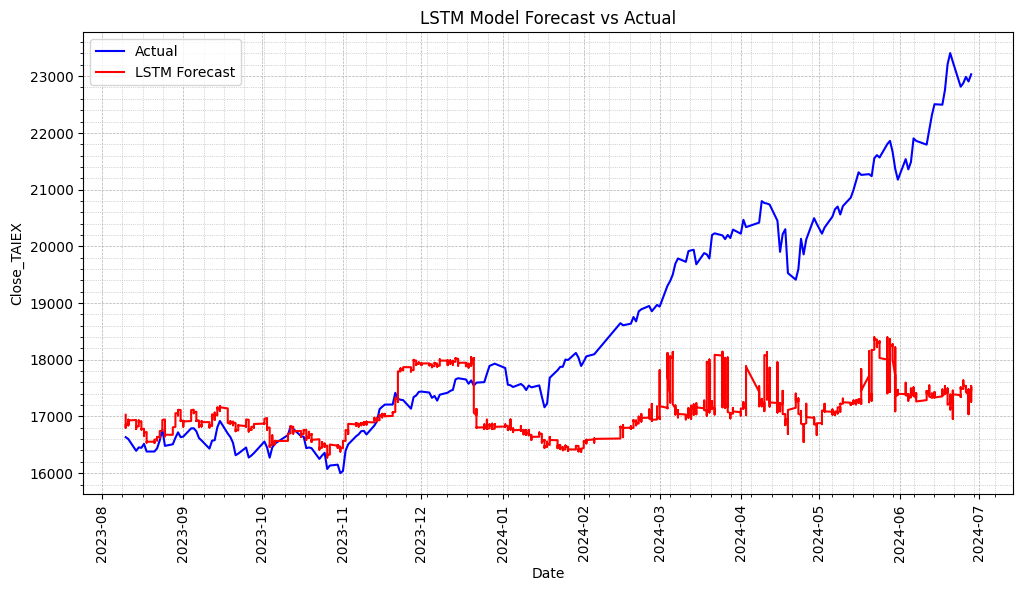
TAIEX.s46_LSTM(3). Future select by Forest
使用決策樹回歸進行特徵選擇,並使用選擇的特徵來訓練 LSTM 模型。最後打印所選的特徵變數並顯示實際值和預測值的圖表。
import os
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
from sklearn.metrics import mean_squared_error, mean_absolute_error
from sklearn.preprocessing import MinMaxScaler
from sklearn.ensemble import RandomForestRegressor
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import LSTM, Dense, Dropout
from google.colab import drive
# Mount Google Drive
drive.mount('/content/drive', force_remount=True)
# Check file path
file_path = '/content/drive/My Drive/MSCI_Taiwan_30_data_with_OBV.csv'
if os.path.exists(file_path):
print("File exists")
data = pd.read_csv(file_path)
else:
print("File does not exist")
# Ensure the date column is converted to datetime type
data['Date'] = pd.to_datetime(data['Date'])
# Print column names to check if 'Close_TAIEX' column exists
print(data.columns)
# Feature Engineering using Adj_Close for moving averages
data['MA7'] = data['Adj_Close'].rolling(window=7).mean()
data['MA21'] = data['Adj_Close'].rolling(window=21).mean()
def compute_rsi(data, window=14):
delta = data['Adj_Close'].diff(1)
gain = (delta.where(delta > 0, 0)).rolling(window=window).mean()
loss = (-delta.where(delta < 0, 0)).rolling(window=window).mean()
rs = gain / loss
rsi = 100 - (100 / (1 + rs))
return rsi
data['RSI14'] = compute_rsi(data)
def compute_macd(data, fast=12, slow=26, signal=9):
exp1 = data['Adj_Close'].ewm(span=fast, adjust=False).mean()
exp2 = data['Adj_Close'].ewm(span=slow, adjust=False).mean()
macd = exp1 - exp2
signal_line = macd.ewm(span=signal, adjust=False).mean()
return macd, signal_line
data['MACD Line'], data['MACD Signal'] = compute_macd(data)
# Prepare data
if 'Close_TAIEX' in data.columns:
data = data.dropna(subset=['Close_TAIEX'])
series = data[['Date', 'Close_TAIEX']]
series.set_index('Date', inplace=True)
else:
print("The data does not contain 'Close_TAIEX' column")
raise KeyError("The data does not contain 'Close_TAIEX' column")
# Selecting Features (excluding non-numeric columns)
numeric_data = data.select_dtypes(include=[np.number])
X = numeric_data.drop(['Close_TAIEX'], axis=1).fillna(0)
y = numeric_data['Close_TAIEX']
# Feature Selection using Random Forest
forest = RandomForestRegressor(n_estimators=100, random_state=42)
forest.fit(X, y)
# Get feature importances
importances = forest.feature_importances_
indices = np.argsort(importances)[::-1]
# Select top 6 features
selected_features = X.columns[indices][:6]
print("Selected features:", selected_features)
# Using the selected features for LSTM model
X_selected = data[selected_features].fillna(0)
# Scale data
scaler_X = MinMaxScaler(feature_range=(0, 1))
scaler_y = MinMaxScaler(feature_range=(0, 1))
scaled_X = scaler_X.fit_transform(X_selected)
scaled_y = scaler_y.fit_transform(y.values.reshape(-1, 1))
# Prepare training and testing datasets
train_size = int(len(scaled_X) * 0.8)
train_X, test_X = scaled_X[:train_size], scaled_X[train_size:]
train_y, test_y = scaled_y[:train_size], scaled_y[train_size:]
# Create sequences
def create_sequences(data_X, data_y, seq_length):
X, y = [], []
for i in range(len(data_X) - seq_length):
X.append(data_X[i:i+seq_length])
y.append(data_y[i+seq_length])
return np.array(X), np.array(y)
seq_length = 60
X_train, y_train = create_sequences(train_X, train_y, seq_length)
X_test, y_test = create_sequences(test_X, test_y, seq_length)
# Build LSTM model
model = Sequential()
model.add(LSTM(units=50, return_sequences=True, input_shape=(seq_length, len(selected_features))))
model.add(Dropout(0.2))
model.add(LSTM(units=50, return_sequences=False))
model.add(Dropout(0.2))
model.add(Dense(units=1))
model.compile(optimizer='adam', loss='mean_squared_error')
model.fit(X_train, y_train, epochs=50, batch_size=32)
# Make predictions
predicted = model.predict(X_test)
predicted = scaler_y.inverse_transform(predicted)
# Calculate errors
y_test_inverse = scaler_y.inverse_transform(y_test)
mse = mean_squared_error(y_test_inverse, predicted)
mae = mean_absolute_error(y_test_inverse, predicted)
# Plot actual values and predictions
plt.figure(figsize=(12, 6))
# Plot actual values
plt.plot(series.index[-len(y_test):], y_test_inverse, label='Actual', color='blue')
# Plot predictions
plt.plot(series.index[-len(predicted):], predicted, label='LSTM Forecast', color='red')
plt.title('LSTM Model Forecast vs Actual')
plt.xlabel('Date')
plt.ylabel('Close_TAIEX')
plt.legend()
# Set date format and rotate x-axis labels
plt.gca().xaxis.set_major_locator(mdates.MonthLocator())
plt.gca().xaxis.set_major_formatter(mdates.DateFormatter('%Y-%m'))
plt.gca().tick_params(axis='x', rotation=90)
# Add grid
plt.grid(True, which='both', linestyle='--', linewidth=0.5)
plt.minorticks_on()
plt.grid(True, which='minor', linestyle=':', linewidth=0.5)
# Show plot
plt.show()
# Print MSE and MAE
print(f'LSTM Model Mean Squared Error (MSE): {mse}')
print(f'LSTM Model Mean Absolute Error (MAE): {mae}')
# Print all selected features for reference
print("Selected features used in the model:", selected_features)
LSTM Model Mean Squared Error (MSE): 5515095.63129834
LSTM Model Mean Absolute Error (MAE): 1712.2863692531828
Selected features used in the model: Index(['OBV', 'Signal Line', 'Market Return', 'MACD Histogram', 'Aroon Down','CCI20'],dtype='object')